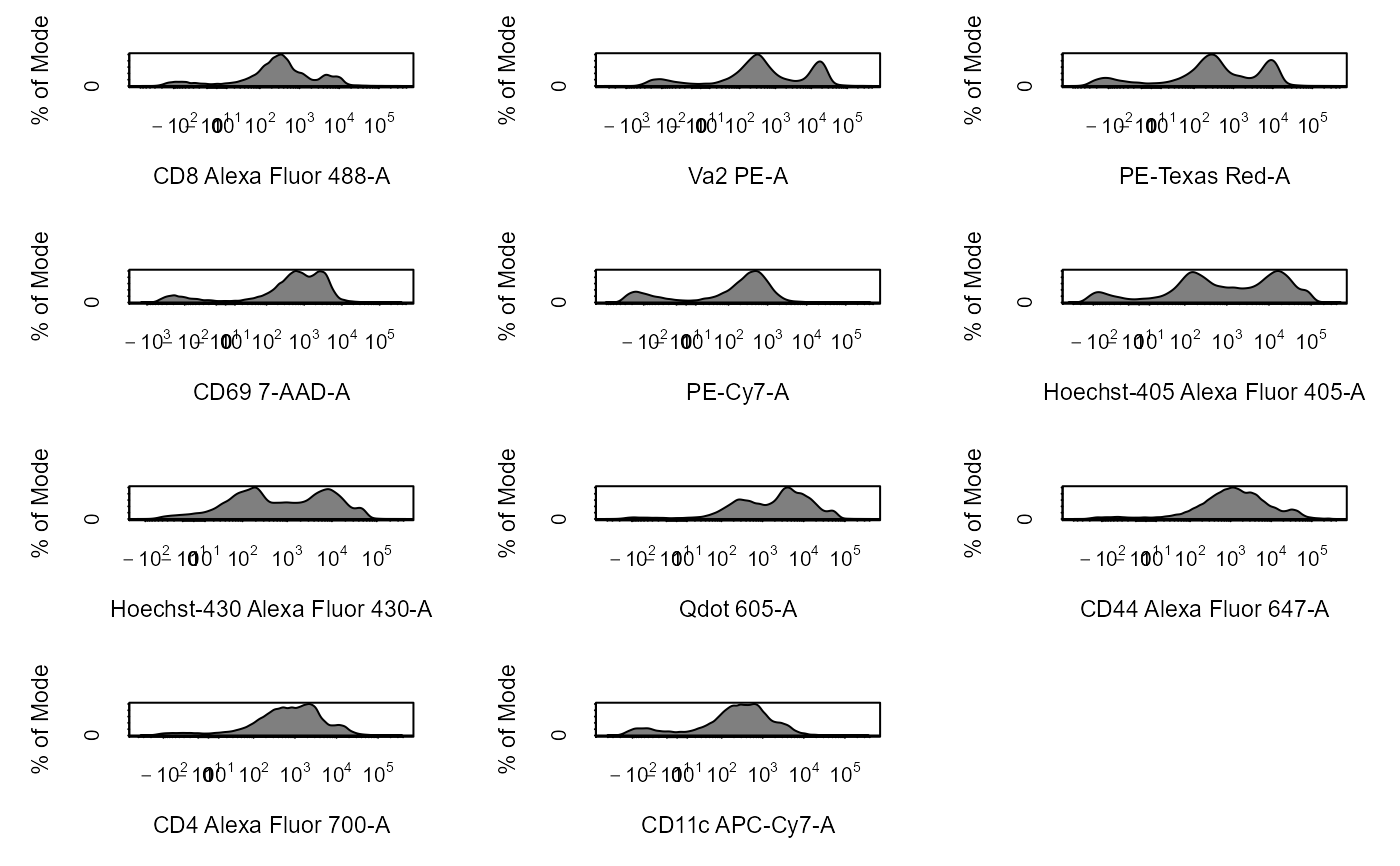
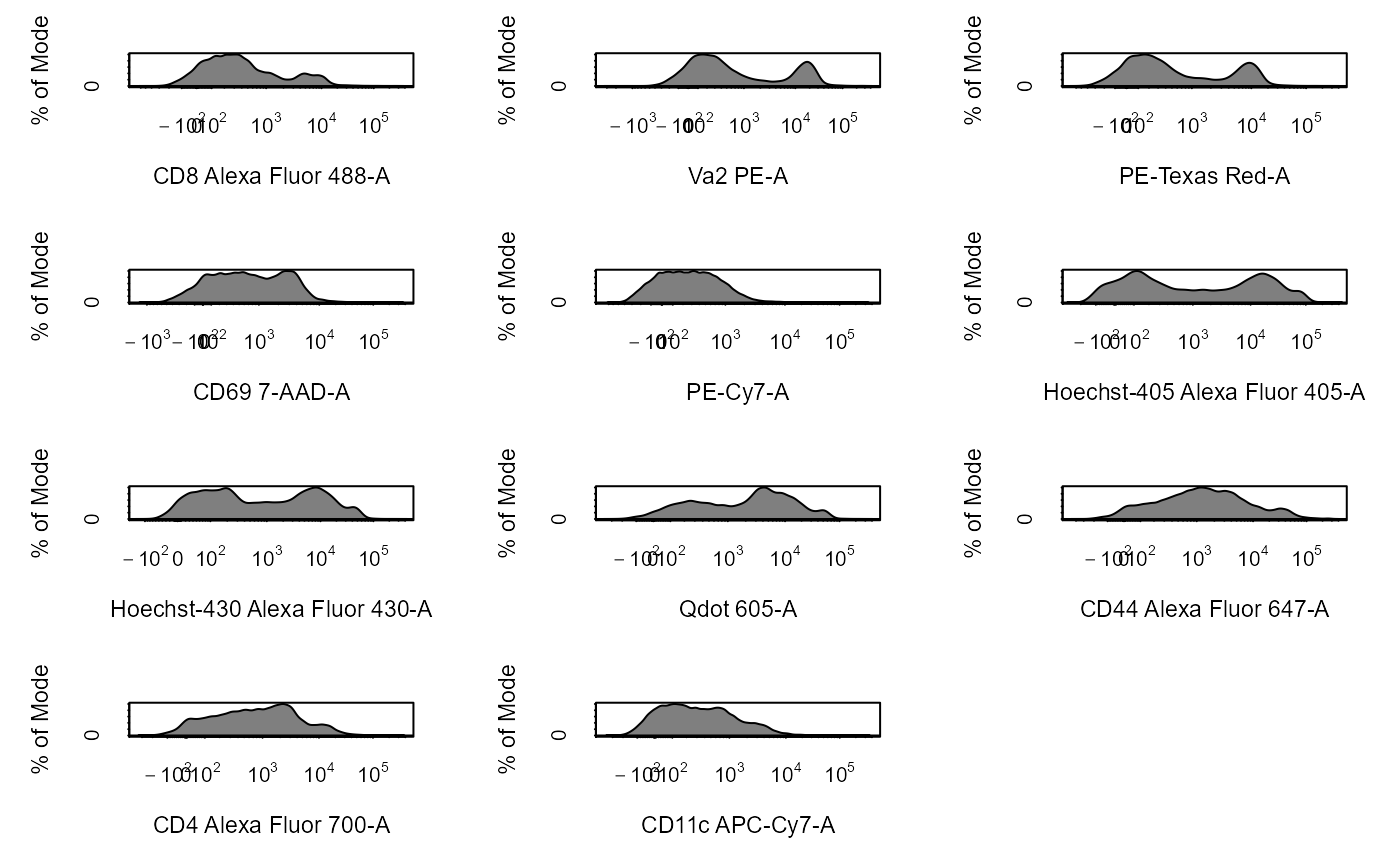
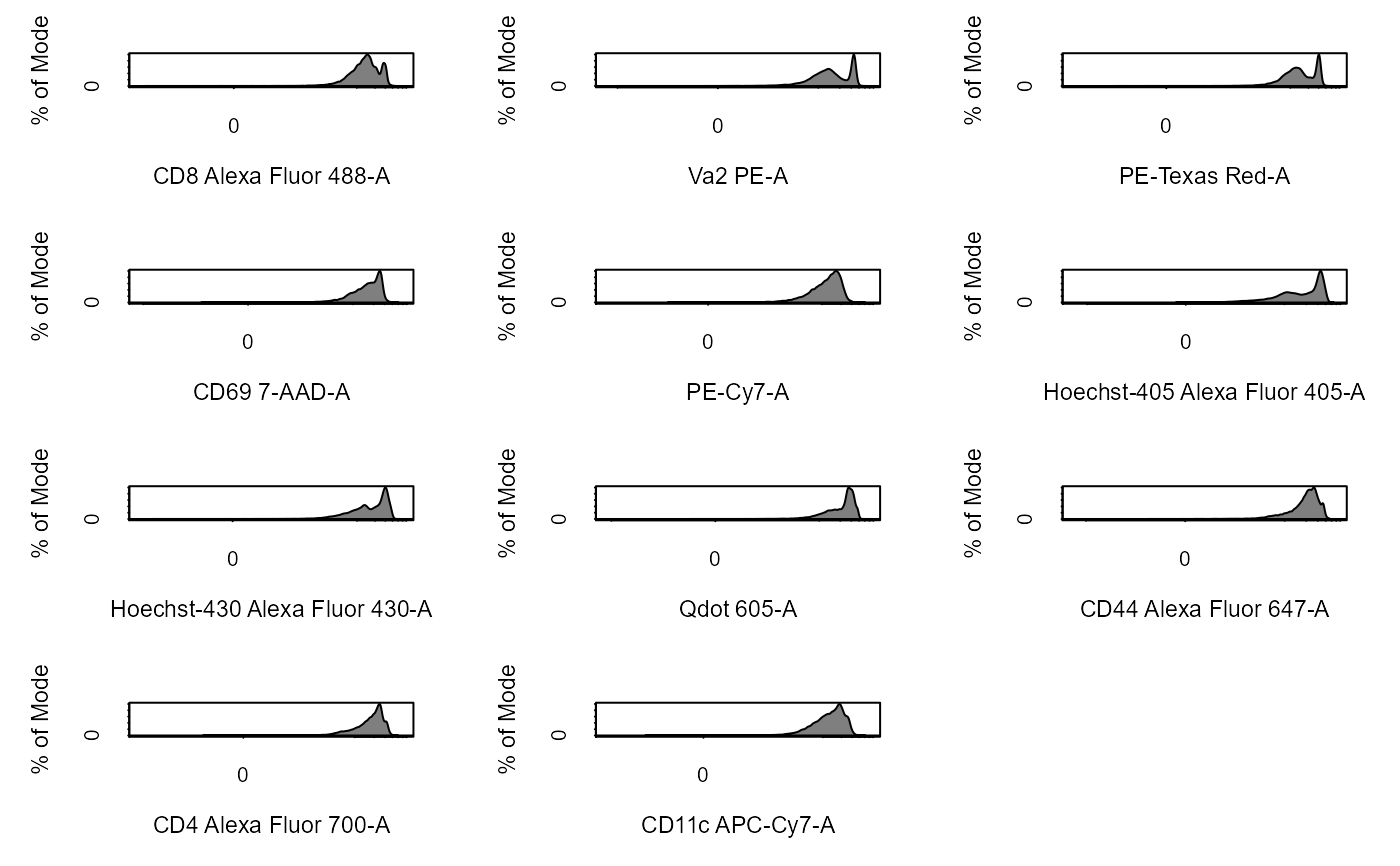
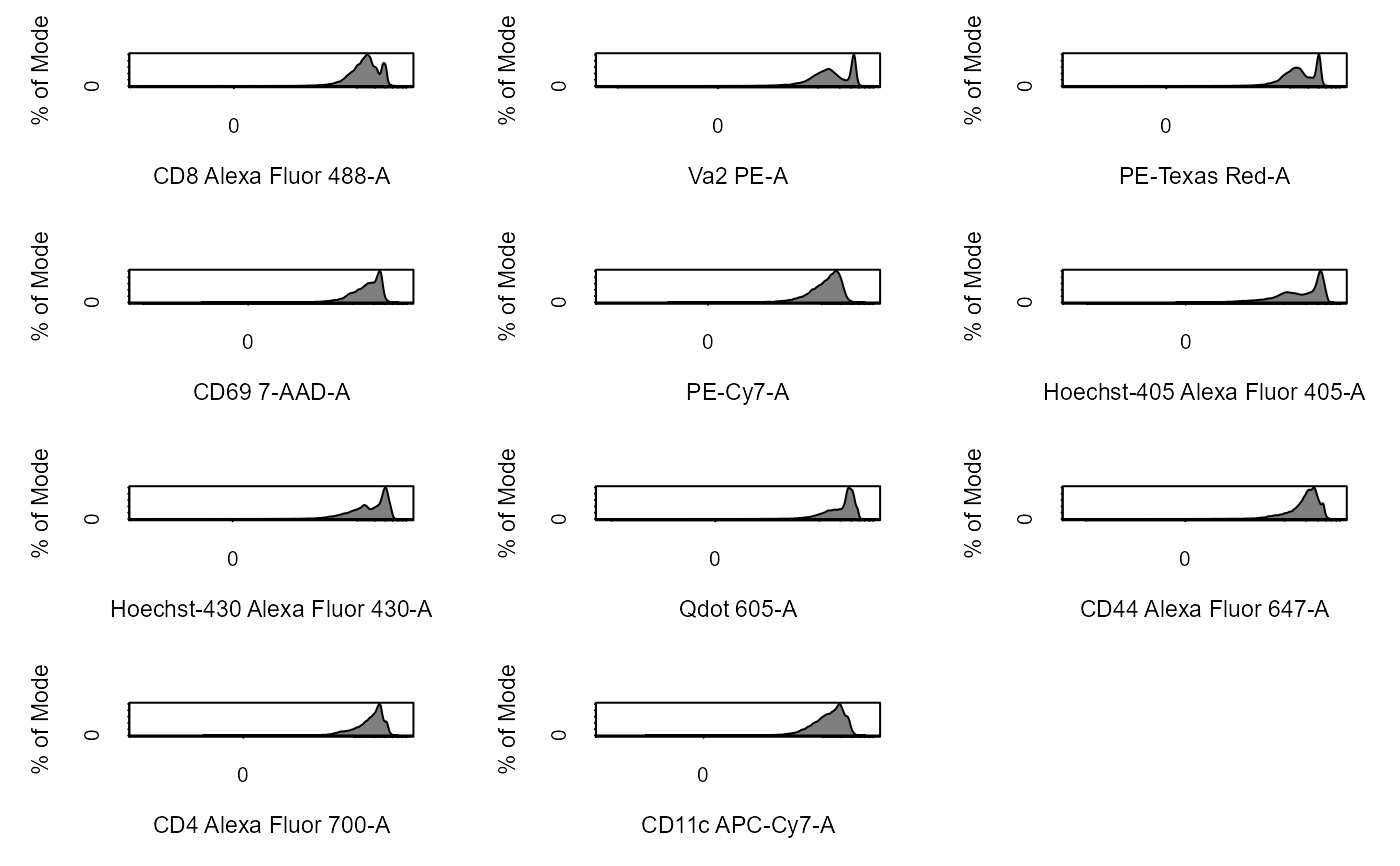
Apply Transformations to Cytometry Data
cyto_transform(x, trans = NULL, ...) # S3 method for default cyto_transform( x, trans = NULL, type = "logicle", channels = NULL, parent = "root", select = NULL, inverse = FALSE, plot = TRUE, popup = FALSE, axes_limits = "machine", ... ) # S3 method for transformList cyto_transform( x, trans = NULL, plot = TRUE, popup = FALSE, axes_limits = "machine", ... ) # S3 method for transformerList cyto_transform( x, trans = NULL, inverse = FALSE, plot = TRUE, popup = FALSE, axes_limits = "machine", ... )
Arguments
| x | object of class |
|---|---|
| trans | object of class
|
| ... | additional arguments passed to |
| type | type of transformation to apply when no trans object is supplied,
options include |
| channels | names of the channels to transform, set to all the
fluorescent channels by default. Only required when no |
| parent | name of the parent population of |
| select | list of selection criteria passed to |
| inverse | logical indicating whether the inverse transformations should
be applied. Currently only supported for |
| plot | logical indicating whether the result of the transformations
should be plotted using |
| popup | logical indicating whether plots should be constructed in a popup window, set to FALSE by default. |
| axes_limits | options include |
Value
object of class flowFrame, flowSet,
GatingHierarchy or GatingSet with transformations applied.
See also
Author
Dillon Hammill, Dillon.Hammill@anu.edu.au
Examples
# Load in CytoExploreRData to access data library(CytoExploreRData) # Activation flowSet fs <- Activation # Automatically transform flowSet fs_trans <- cyto_transform(fs, type = "arcsinh")#>fs_trans <- cyto_transform(fs, trans)# Add fs to GatingSet gs <- GatingSet(fs) # Automatically transform GatingSet gs_trans <- cyto_transform(gs, type = "logicle")gs_trans <- cyto_transform(gs, trans)