CytoExploreR is comprehensive collection of interactive exploratory cytometry analysis tools designed under a unified framework. CytoExploreR has been specifically designed to integrate all existing cytometry analysis techniques (e.g. manual gating, automated gating and dimension reduction) in a format that makes these tools freely accessible to users with no coding experience. If you are new to CytoExploreR visit https://dillonhammill.github.io/CytoExploreR/ to get started.
Install R and RStudio
CytoExploreR has a minimal requirement for R 3.5.0. If necessary, newer versions of R can be installed by clicking on your operating system in this link and following the installation instructions. For the best user experience it is recommended that RStudio be installed as well. RStudio Desktop is free to download and can installed from the RStudio website.
Platform-Specific Requirements
After successfully installing R and RStudio, the following additional platform-specific tools are required:
Mac OS
- Install Xcode developer tools from the App Store. Restart your computer.
- Install command line tools by opening the terminal and running xcode-select –install
- Install macOS R toolchain by installing clang7 and gfortran here
- Check if XQuartz is listed in your installed Applications, it may be hiding in the utilities folder. If XQuartz is missing on your computer, it can be installed from the XQuartz website.
- Restart your computer so all these changes will take effect.
Windows OS
- Install the appropriate Rtools for your R installation.
- Follow these instructions to download protobuf Windows binary and set appropriate environment variable. Ignore the section to build protobuf from source.
- Follow these instructions to install and setup the additional C++ libraries required to successfully build flowWorkspace.
- Restart your computer.
CytoExploreR Dependencies
Now that all the setup is complete, let’s install all the necessary dependencies of CytoExploreR. From within RStudio, run the following in the console to install the latest versions of the cytoverse packages from Bioconductor.
# Bioconductor
install.packages("BiocManager")
# Install cytoinstaller
remotes::install_github("RGLab/cytoinstaller")
# Install cytoverse packages
cytoinstaller::install_cyto(bioc_ver = "devel")CytoExploreR
Now that all the dependencies are installed, let’s move on to installing CytoExploreR. To successfully install CytoExploreR users will first need to install CytoExploreRData which contains example datasets that will be used within CytoExploreR to demonstrate key features.
# CytoExploreRData
devtools::install_github("DillonHammill/CytoExploreRData")
# CytoExploreR
devtools::install_github("DillonHammill/CytoExploreR")Design
To ease the transition from GUI oriented software, CytoExploreR has been designed to be a consistent and auto-complete friendly package for cytometry data analysis. All exported functions from CytoExploreR are prefixed with cyto_, followed by the name of the object you wish to change (e.g. cyto_gate_) and finally the action that you would like to perform (e.g. cyto_gate_draw). To see all available functions simply start typing cyto_ and you will be greeted with a complete list of exported functions that can be selected from the auto-complete dropdown list:
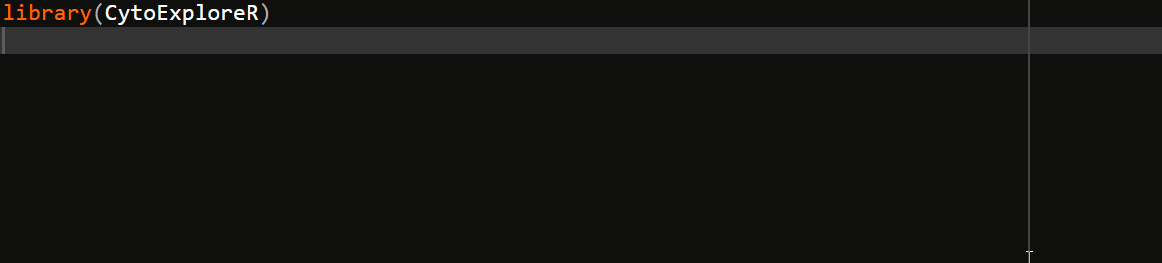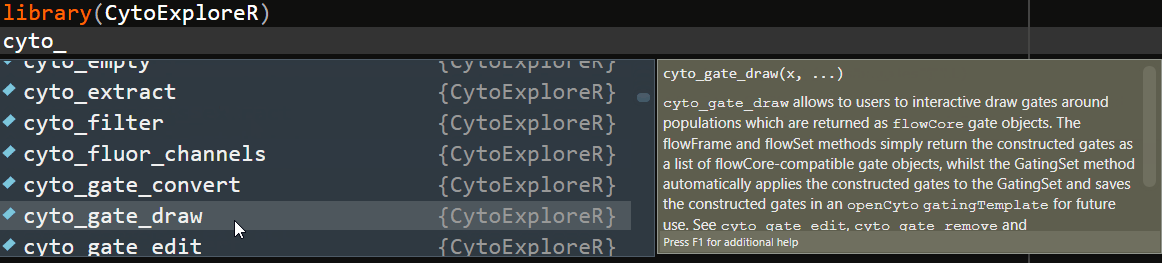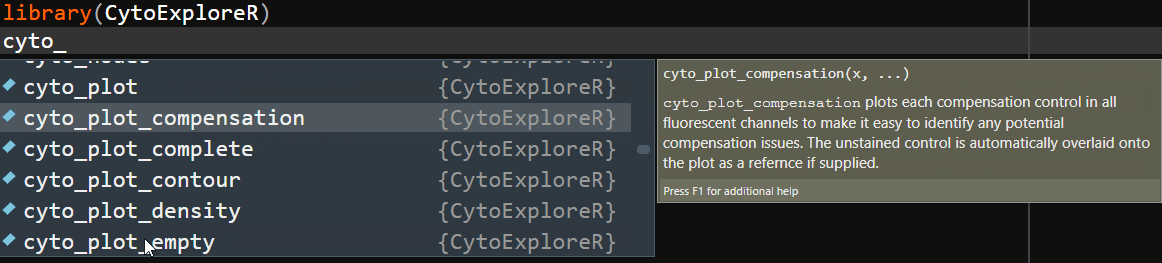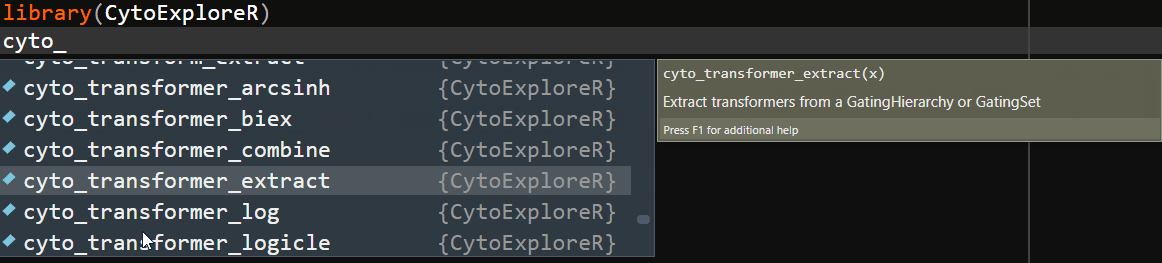
Overview
Some of the key features of CytoExploreR are outlined below:
- load and annotate samples using
cyto_setup - user guided automatic compensation using
cyto_spillover_compute - interactively modify spillover matrices using
cyto_spillover_edit - compute spillover spreading matrices with
cyto_spillover_spread_compute - visualise compensation in all channels using
cyto_plot_compensation - customisable data transformations using
cyto_transformwhich includes support for log, arcsinh, logicle and biexponential data transformations - manual gate drawing using
cyto_gate_draw - ability to edit drawn gates using
cyto_gate_edit - remove gates using
cyto_gate_remove - rename gates using
cyto_gate_rename - gate saving directly to an openCyto
gatingTemplatefor future use - support for using both manual and automated gating approaches through linking to
openCyto - exploratory visualisations of all existing cytometry data classes using
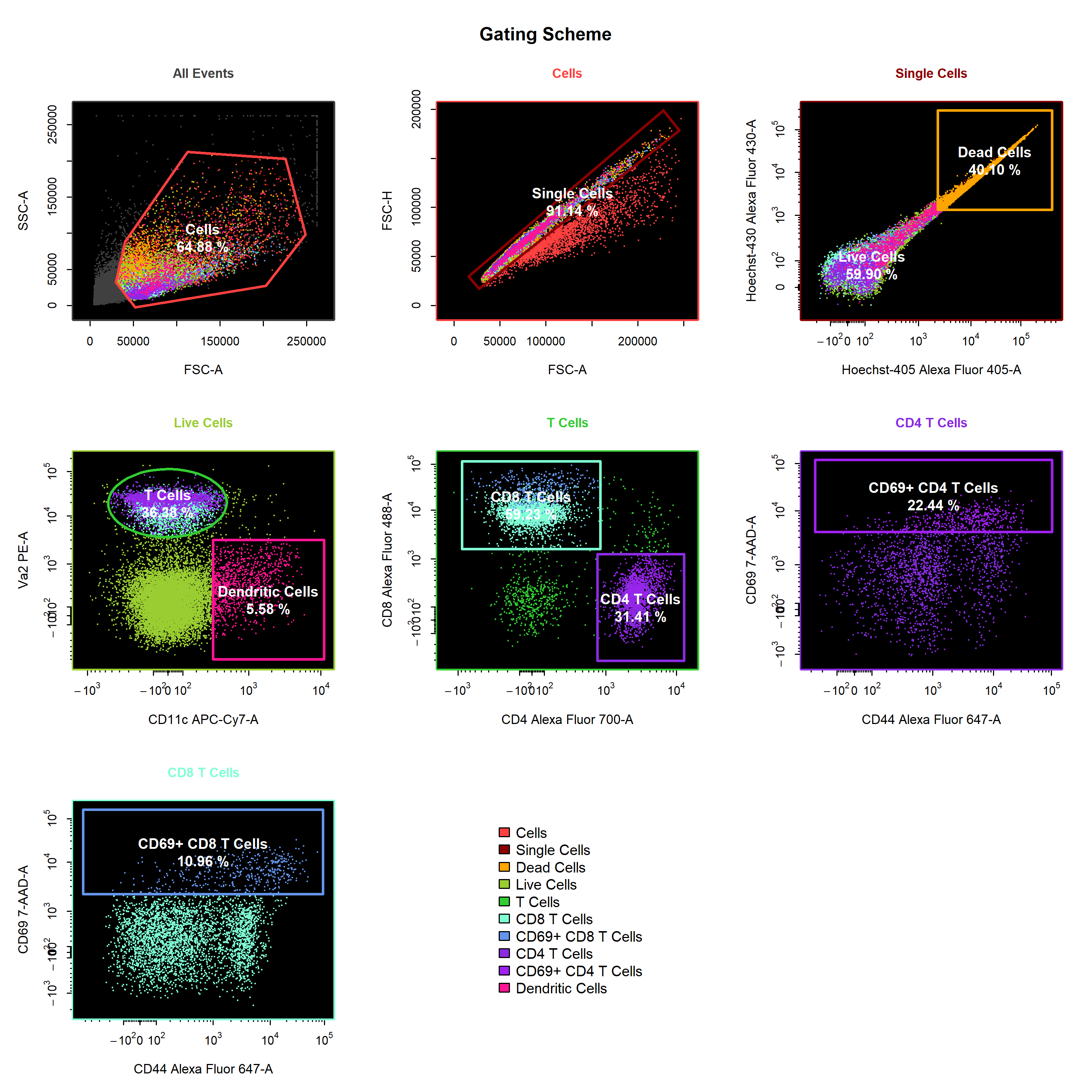
cyto_plot - visualisation of complete gating strategies with back-gating and/or gate tracking using
cyto_plot_gating_scheme - visualisation of gating trees using
cyto_plot_gating_tree - visualisation of marker expression profiles in all channels using
cyto_plot_profile - visualisation of populations in all possible bivariate plots using
cyto_plot_explore - produce dimension reduced maps (e.g. PCA, tSNE, UMAP and EmbedSOM) using
cyto_map - save samples and analyses to file using
cyto_save - export population level statistics tidyverse style using
cyto_stats_compute
Usage
CytoExploreR is large package and we would not do it justice by demonstrating its usage here. Instead we will explore the use of CytoExploreR in a series of vignettes which tackle specific components of the cytometry data analysis pipeline. To work through these vignettes you will need to create a new R project (File -> New Project) and download the example datasets shipped with CytoExploreRData.
# Load required packages
library(CytoExploreR)
library(CytoExploreRData)
# Download Compensation FCS files
cyto_save(Compensation,
save_as = "Compensation-Samples")
# Download Activation FCS files
cyto_save(Activation,
save_as = "Activation-Samples")These datasets will be used throughout the package vignettes to demonstrate the key features of CytoExploreR. A brief summary of each of the package vignettes is provided below:
The
CytoExploreRvignette outlines a basic flow cytometry data analysis pipeline, which includes steps to compensate for fluorescent spillover, transform data for visualisation and manually gate populations to export population level statistics. This vignette serves as a basic introduction to the package and users are encouraged to explore other vignettes which explore these aspects in a lot more detail.The
Visualisationsvignette will demonstrate the use ofcyto_plot, a powerful data visualisation tool to explore cytometry data.
- Flow cytometry users will find the
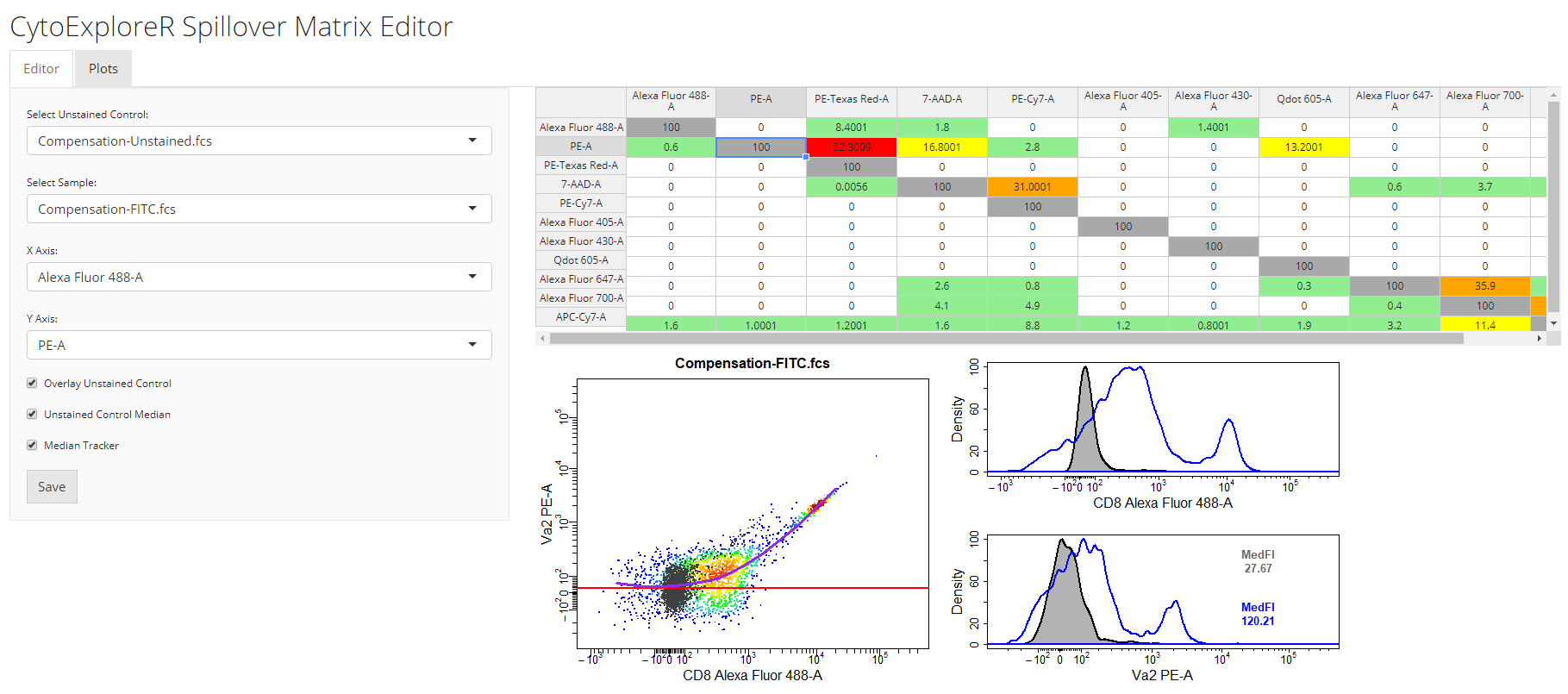
Compensationvignette useful in describing the process of using compensation controls to correctly compensate for fluorescent spillover.
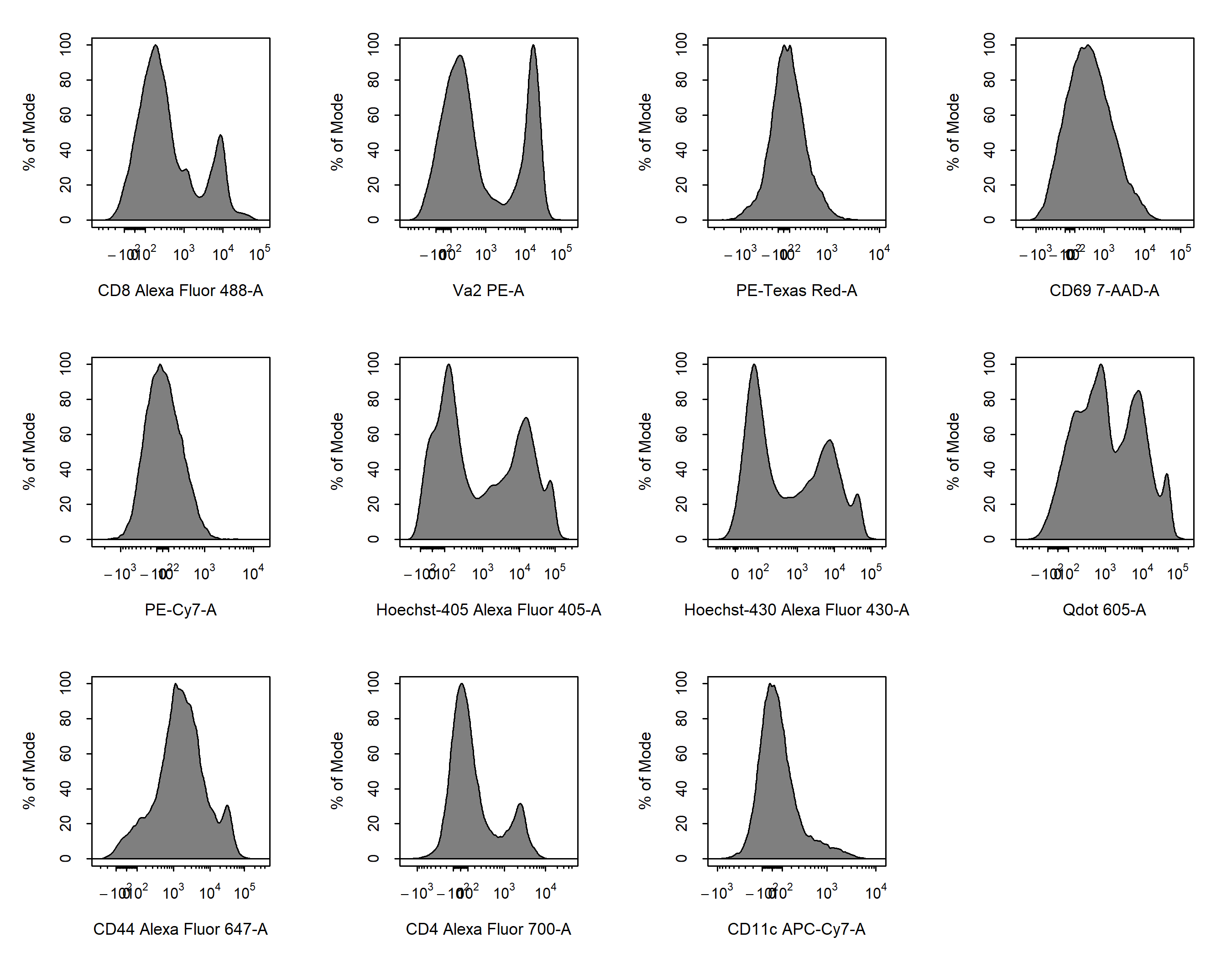
- Data transformations are essential to appropriate visualisation of cytometry data. In the
Transformationsvignette we will explore the tools available in CytoExploreR to apply log, arcsinh, biexponential and/or logicle transformations to the data.
- In
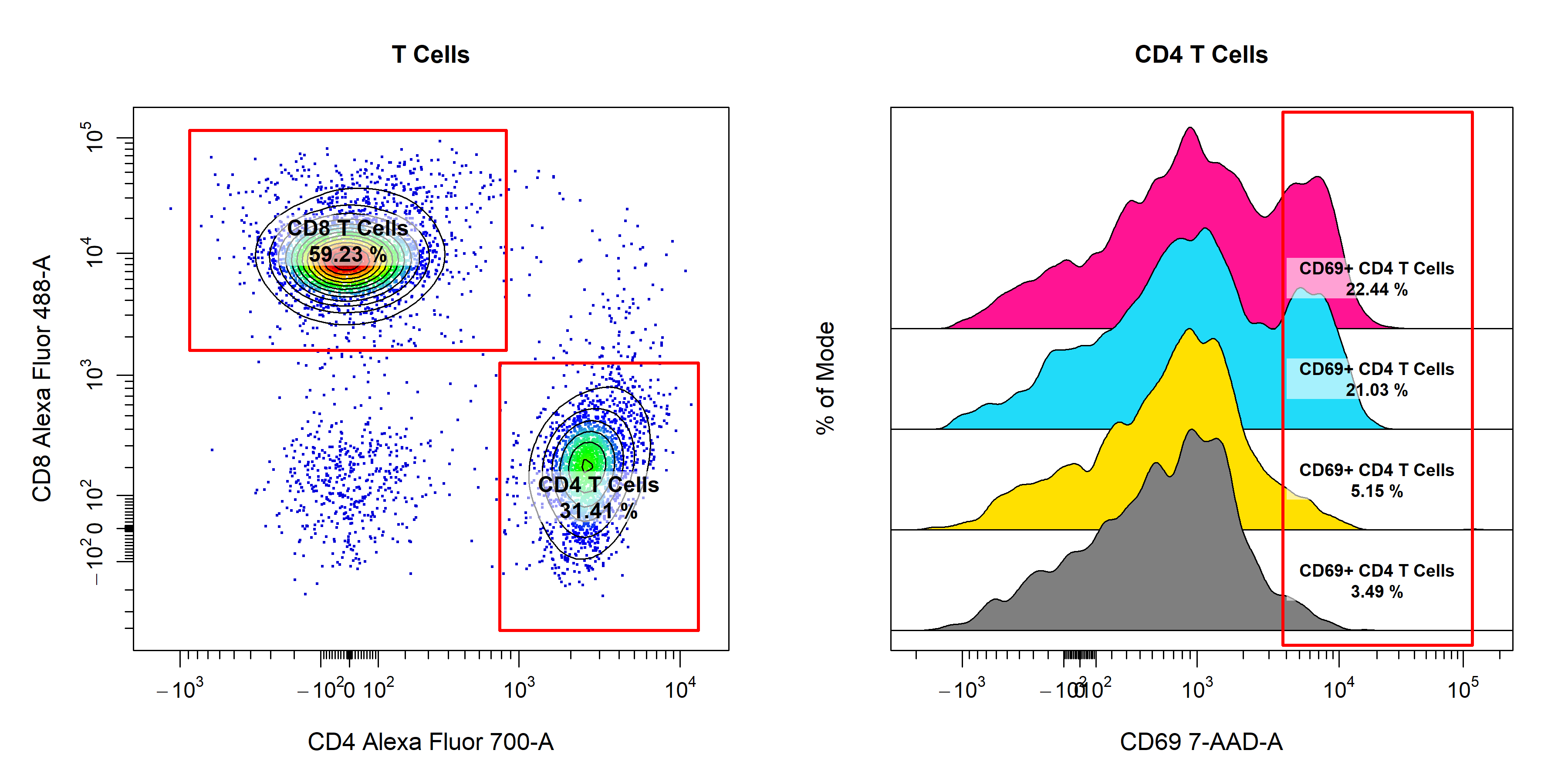
Manual Gatingvignette we will demonstrate the use ofcyto_gate_drawto interactively draw gates around populations. In particular, we will focus on the different gate types that are supported and how they can be used to gate populations.
- Dimensionality reduction is gaining popularity for analysis of high dimensional cytometry data. In the
Dimensionality Reductionvignette we will demonstrate the use ofcyto_mapto produce PCA, tSNE, FIt-SNE, UMAP and EmbedSOM maps of cytometry data (coming soon).
News
There is a changelog for the GitHub master branch which will reflect any updates made to improve the stability, usability or plenitude of the package. Users should refer to the Changelog prior to installing new versions of the package.
Credits
CytoExploreR would not be possible without the existing flow cytometry infrastructure developed by the RGLab. CytoExploreR started out as simple plugin for openCyto to facilitate gate drawing but has evolved into a fully-fledged cytometry analysis package thanks to the support and guidance of members of the RGLab. Please take the time to check out their work on GitHub.
Development
CytoExploreR is a maturing package which will continue to be sculpted by the feedback and feature requests of users. The GitHub master branch will always contain the most stable build of the package. New features and updates will be made to a separate branch and merged to the master branch when stable and tested. The Changelog will reflect any changes made to the master branch.
Getting help
The Get Started and Reference sections on the CytoExploreR website are your first port of call if you require any help. For more detailed workflows refer the Articles tab. If you encounter any issues with the functioning of the package refer to these issues to see if the problem has been identified and resolved. Feel free to post new issues on the GitHub page if they have not already been addressed.
Code of conduct
Please note that the CytoExploreR project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Citation
A CytoExploreR publication is on the way, but in the meantime if you use CytoExploreR for your work please cite the package as follows:
citation("CytoExploreR")
#>
#> To cite package 'CytoExploreR' in publications use:
#>
#> Dillon Hammill (2021). CytoExploreR: Interactive Analysis of
#> Cytometry Data. R package version 1.1.0.
#> https://github.com/DillonHammill/CytoExploreR
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {CytoExploreR: Interactive Analysis of Cytometry Data},
#> author = {Dillon Hammill},
#> year = {2021},
#> note = {R package version 1.1.0},
#> url = {https://github.com/DillonHammill/CytoExploreR},
#> }

