Add contour lines to cyto_plot
cyto_plot_contour(x, ...) # S3 method for flowFrame cyto_plot_contour( x, channels, contour_lines = 15, contour_line_type = 1, contour_line_width = 1, contour_line_col = "black", contour_line_alpha = 1, ... ) # S3 method for flowSet cyto_plot_contour( x, channels, contour_lines = 15, contour_line_type = 1, contour_line_width = 1, contour_line_col = "black", contour_line_alpha = 1, ... ) # S3 method for list cyto_plot_contour( x, channels, contour_lines = 15, contour_line_type = 1, contour_line_width = 1, contour_line_col = "black", contour_line_alpha = 1, ... )
Arguments
| x | either an object of class
|
|---|---|
| ... | not in use. |
| channels | channels used to construct the existing cyto_plot. |
| contour_lines | numeric indicating the number of levels to use for contour lines, set to 15 by default. |
| contour_line_type | type of line to use for contour lines, set to 1 by default. |
| contour_line_width | line width for contour lines, set to 1 by default. |
| contour_line_col | colour to use for contour lines, set to "black" by default. |
| contour_line_alpha | numeric [0,1] to control transparency of contour lines, set to 1 by default to remove transparency. |
Author
Dillon Hammill, Dillon.Hammill@anu.edu.au
Examples
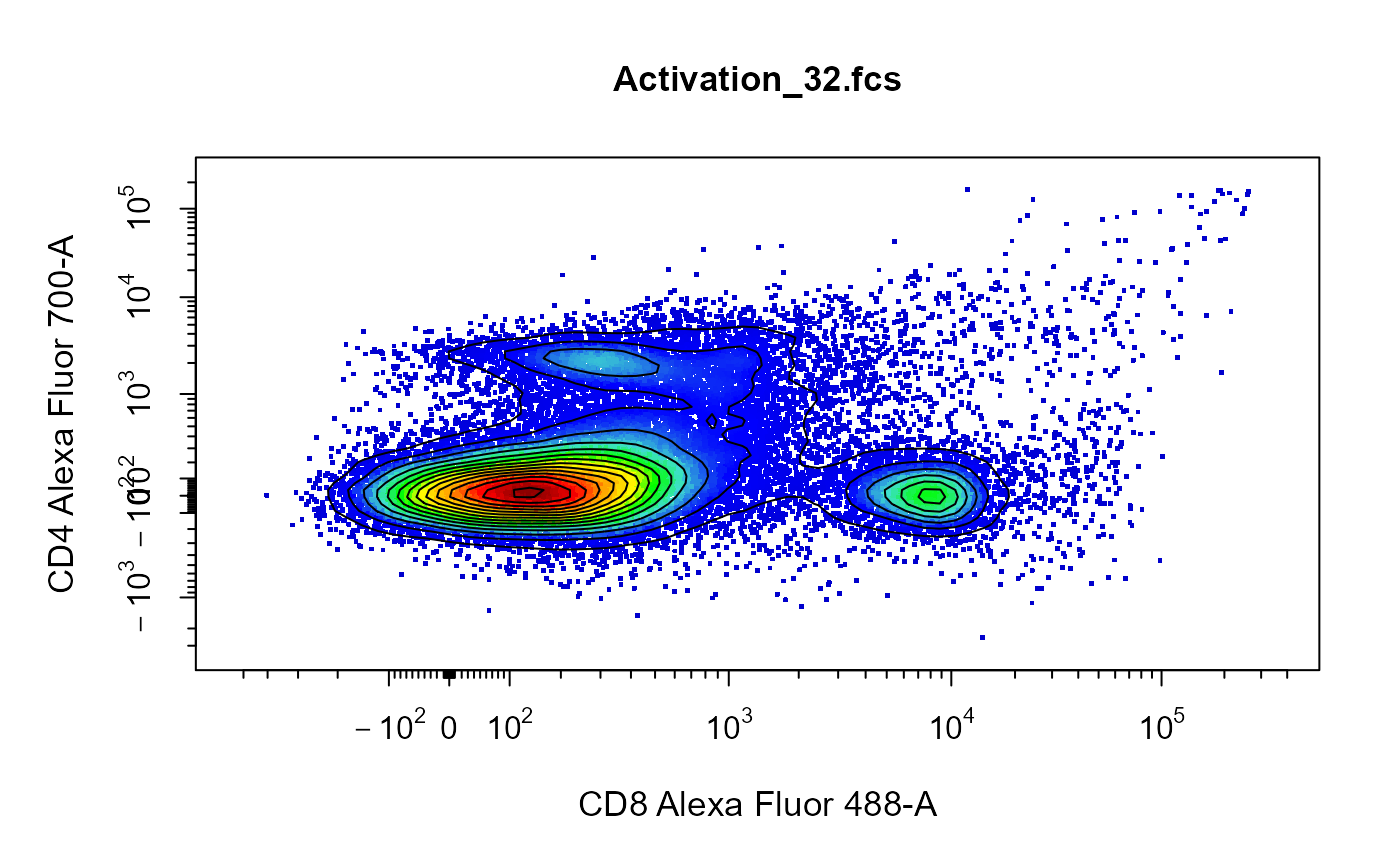
library(CytoExploreRData) # Load in Samples fs <- Activation # Apply compensation fs <- cyto_compensate(fs) # Transform fluorescent channels trans <- cyto_transformer_logicle(fs)# Plot cyto_plot(fs[[32]], channels = c("Alexa Fluor 488-A", "Alexa Fluor 700-A"), axes_trans = trans )# Contour lines cyto_plot_contour(fs[[32]], channels = c("Alexa Fluor 488-A", "Alexa Fluor 700-A"), contour_lines = 20 )