Compute Spillover Matrix - GatingSet Method
Source:R/spillover_compute-methods.R
spillover_compute-GatingSet-method.Rdspillover_compute uses the method described by Bagwell & Adams 1993 to
calculate the fluorescent spillover matrix using a reference universal
unstained control and single stain compensation controls.
# S4 method for GatingSet spillover_compute(x, parent = NULL, axes_trans = NULL, channel_match = NULL, spillover = "Spillover-Matrix.csv", ...)
Arguments
| x | object of class
|
|---|---|
| parent | name of the pre-gated population to use for downstream calculations, set to the last node of the GatingSet by default (e.g. "Single Cells"). |
| axes_trans | object of class
|
| channel_match | name of .csv file containing the names of the samples in
a column called "name" and their matching channel in a column called
"channel". |
| spillover | name of the output spillover csv file, set to
|
| ... | additional arguments passed to
|
Value
spillover matrix object and "Spillover Matrix.csv" file.
Details
Calculate spillover matrix using
GatingSet containing gated
single stain compensation controls and an unstained control.
spillover_compute uses the method described by Bagwell & Adams 1993 to
calculate fluorescent spillover values using single stain compensation
controls and a universal unstained control. spillover_compute begins
by the user selecting which fluorescent channel is associated with each
control from a dropdown menu. Following channel selection,
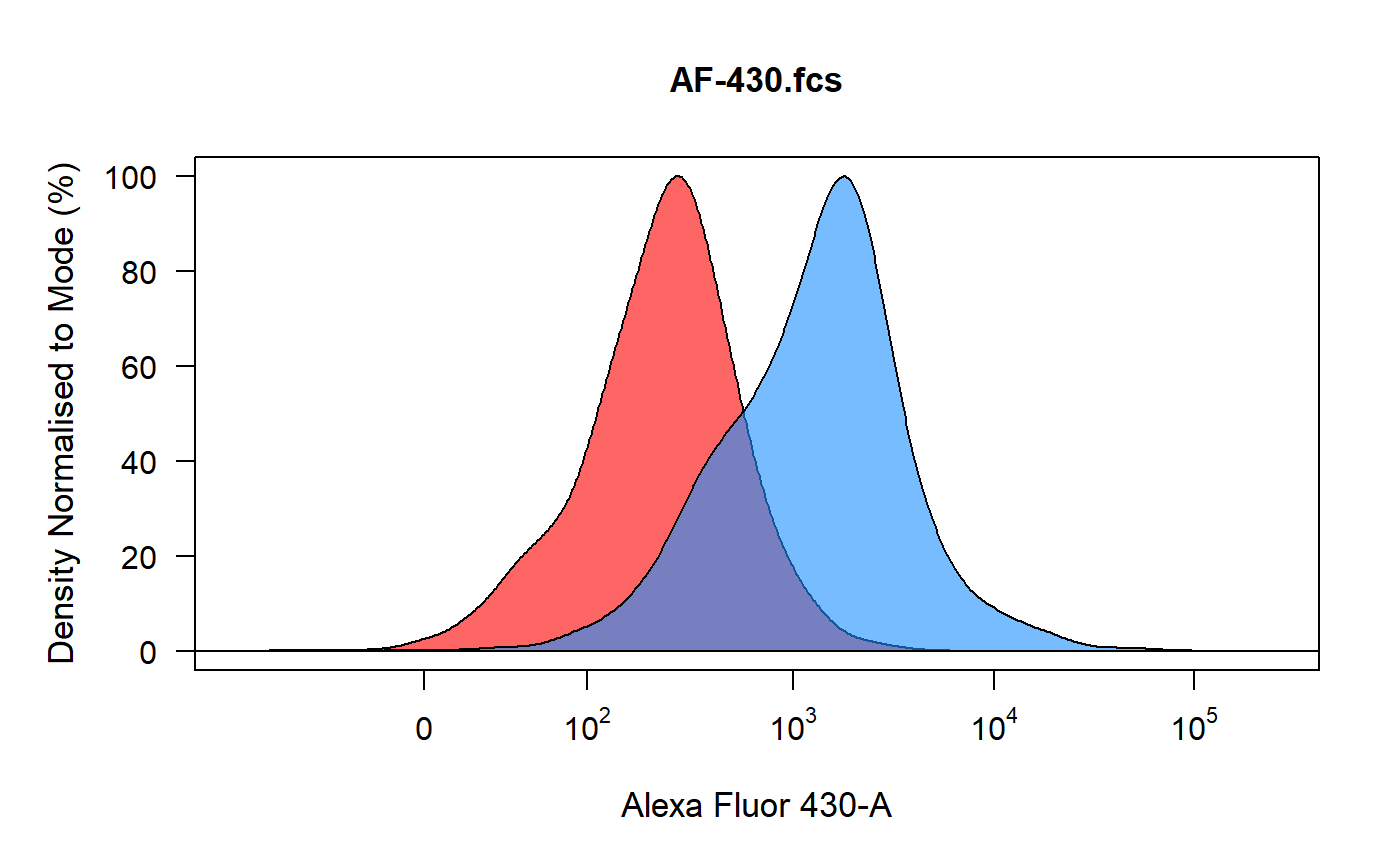
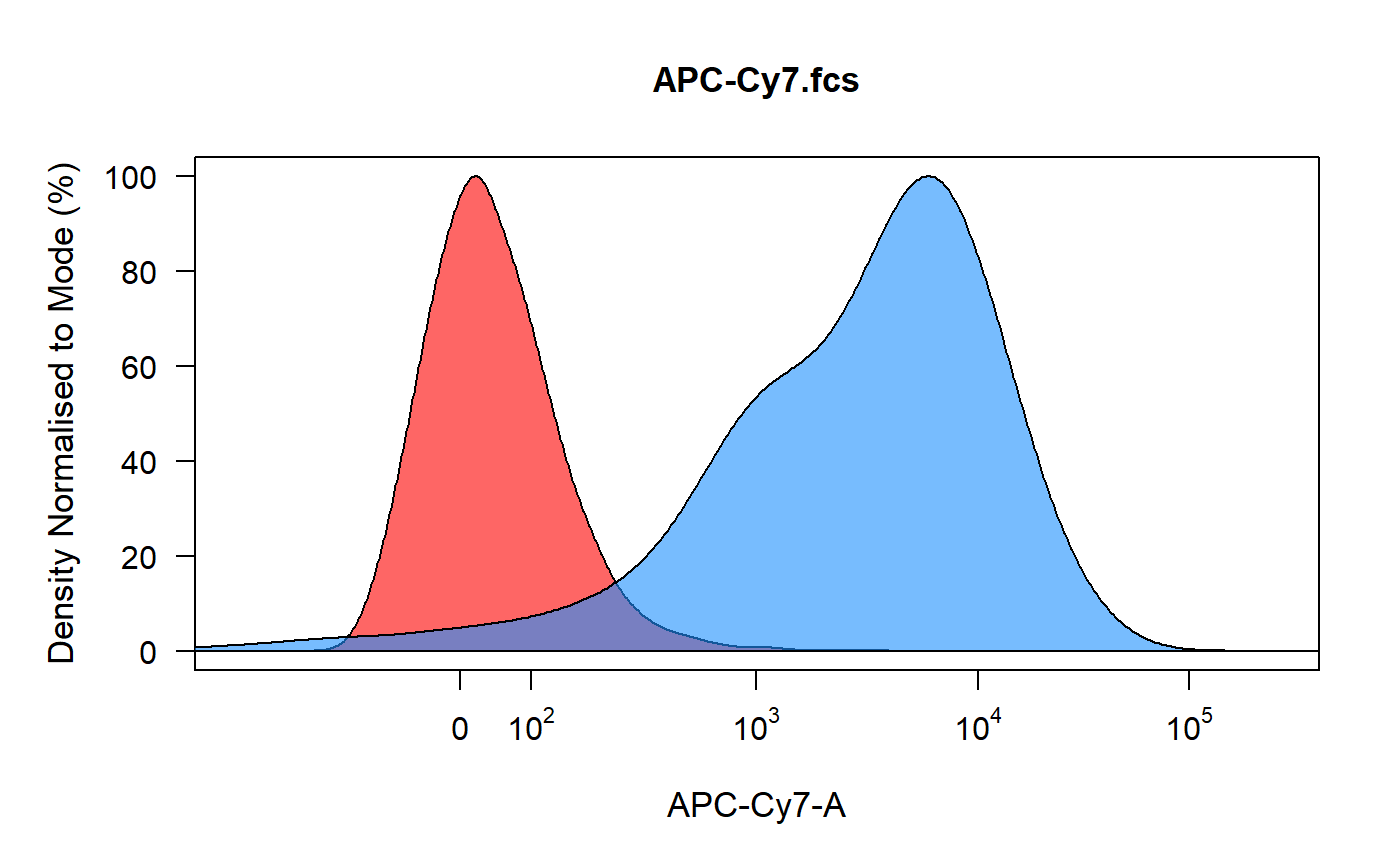
spillover_compute runs through each control and plots the density
distribution of the unstained control in red and the compensation control in
blue. Users can then gate the positive signal for spillover calculation using
an interval gate. The percentage spillover is calculated based on the median
fluorescent intensities of the stained populations and the universal
unstained sample. The computed spillover matrix is returned as an R object
and written to a named .csv file for future use.
References
C. B. Bagwell \& E. G. Adams (1993). Fluorescence spectral overlap compensation for any number of flow cytometry parameters. in: Annals of the New York Academy of Sciences, 677:167-184.
See also
Examples
library(CytoRSuiteData) # Bypass directory check for external files options("CytoRSuite_wd_check" = FALSE) # Don't run - skips the gating process options("CytoRSuite_interact" = FALSE) # Load in compensation controls fs <- Compensation gs <- GatingSet(Compensation)#>#>#>#>#>#>#># Gate using gate_draw gt <- Compensation_gatingTemplate gating(gt, gs)#>#>#>#>#>#>#># Channel match fille cmfile <- system.file("extdata", "Compensation-Channels.csv", package = "CytoRSuiteData" ) # Compute fluorescent spillover matrix spill <- spillover_compute(gs, parent = "Single Cells", channel_match = cmfile, spillover = "Example-spillover.csv" )