Plot Compensation in All Fluorescent Channels - flowSet Method
Source:R/cyto_plot_compensation-methods.R
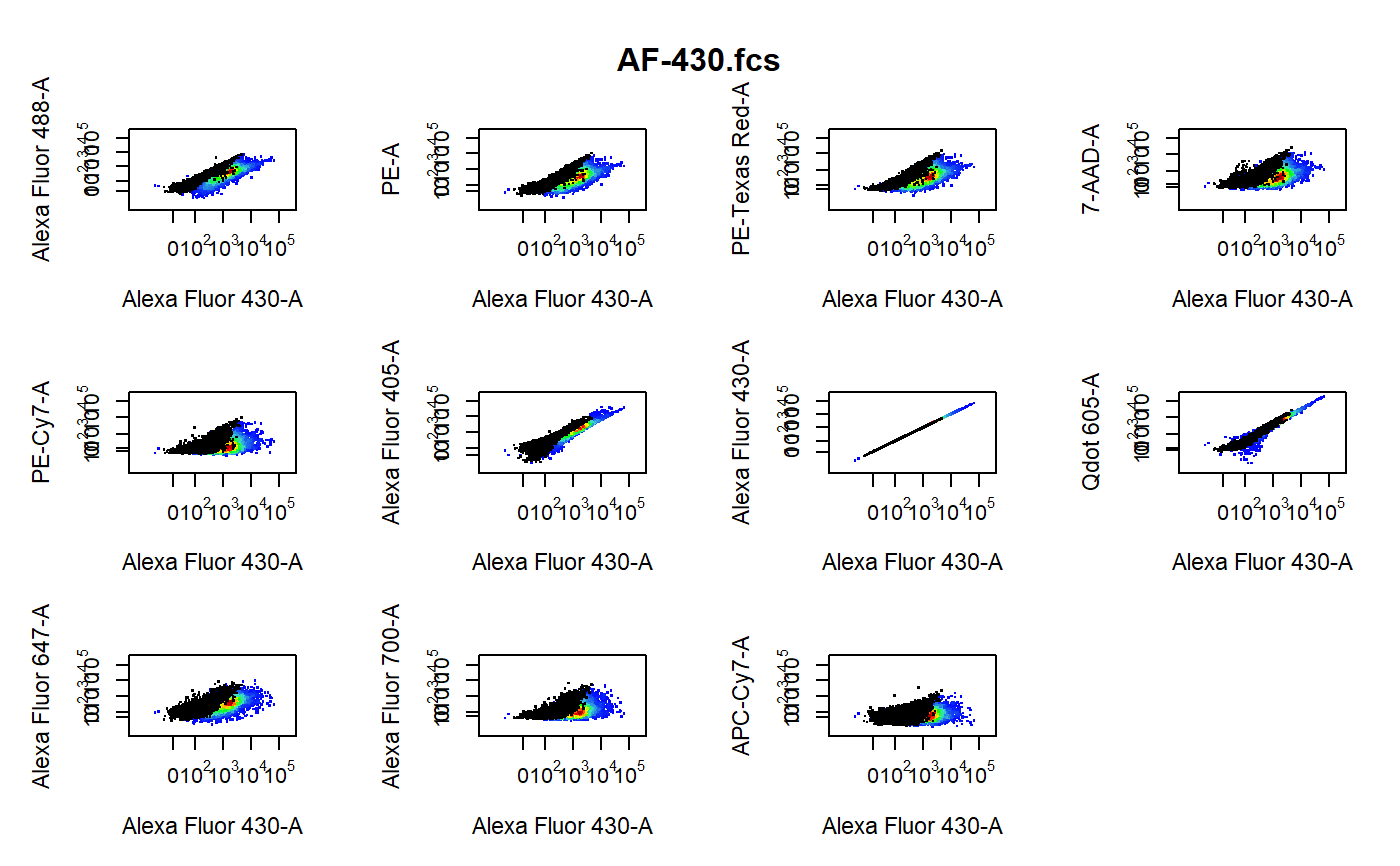
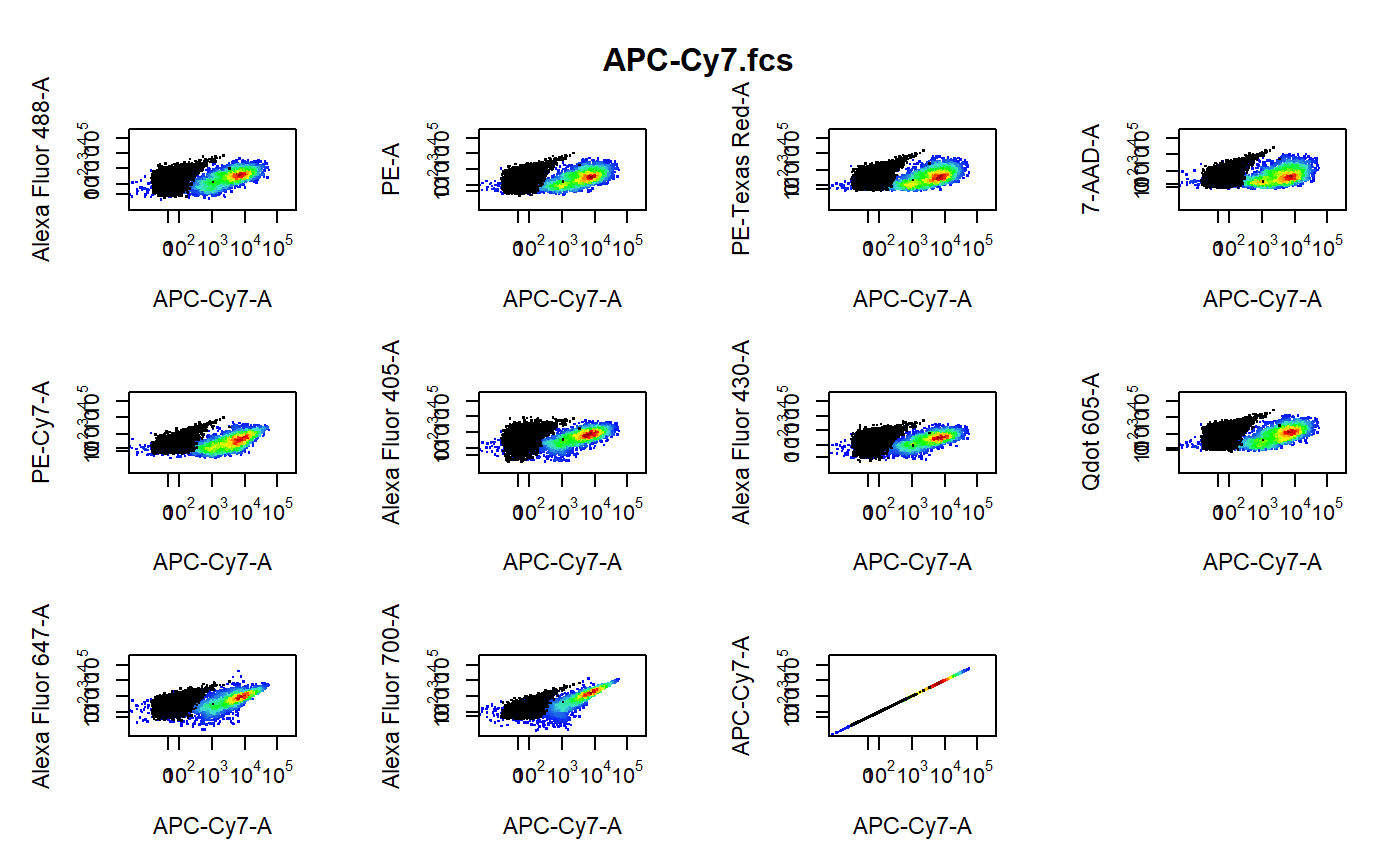
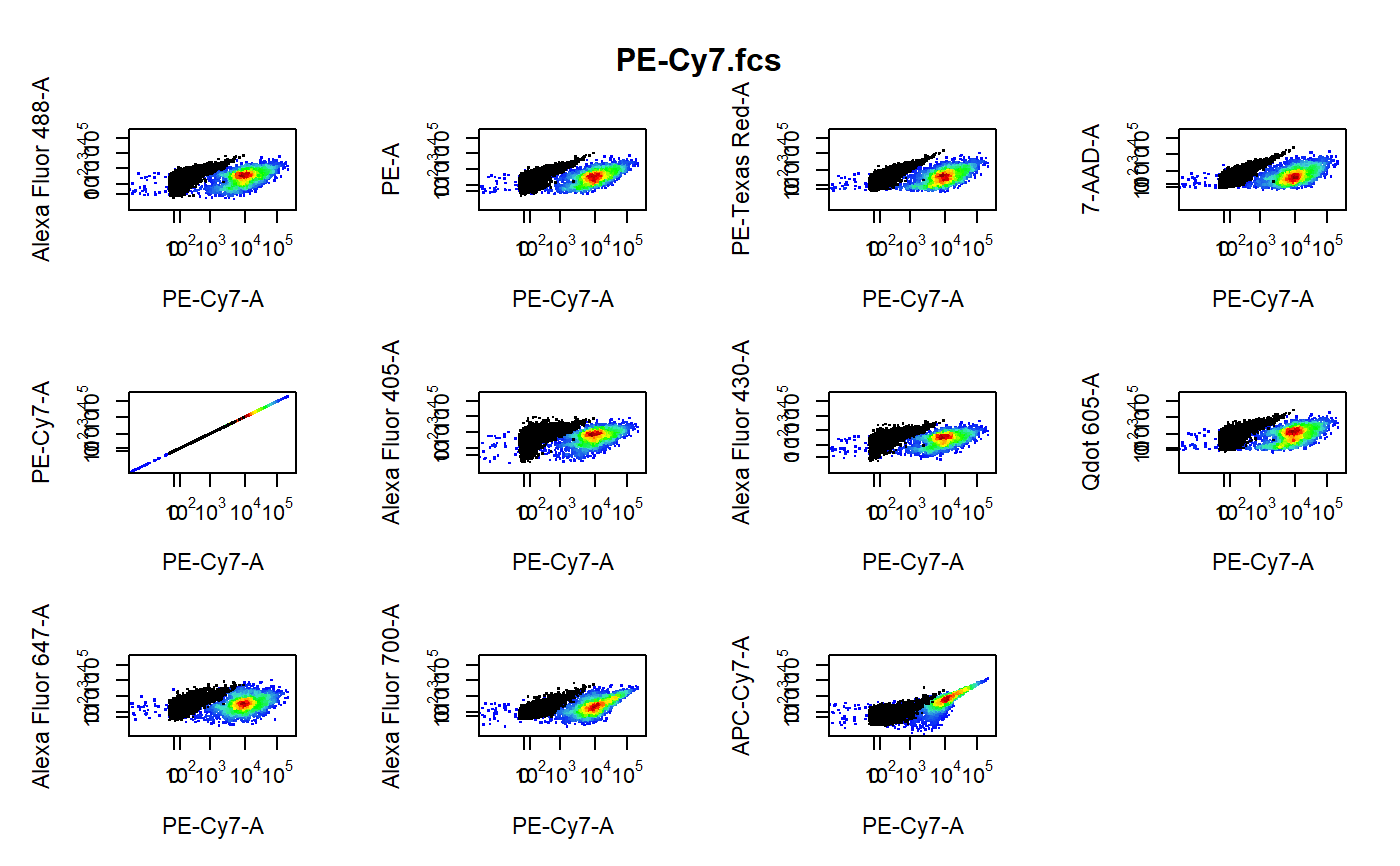
cyto_plot_compensation-flowSet-method.RdPlot each compensation control in all fluorescent channels to identify any potential compensation issues. The unstained control is overlaid in black as a reference.
# S4 method for flowSet cyto_plot_compensation(x, channel_match = NULL, compensate = FALSE, spillover = NULL, axes_trans = NULL, overlay = TRUE, layout, popup = FALSE, title = NA, header = NA, header_text_font = 2, header_text_size = 1, header_text_col = "black", ...)
Arguments
| x | object of class |
|---|---|
| channel_match | name of a csv file with two columns, the first
called "name" lists the names of each compensation control and the second
"channel" lists the fluorescent channel associated with each of the
compensation controls. Use "Unstained" in the channel column for the
universal unstained control. No need to construct this file manually as
users will be guided through this process if the |
| compensate | logical indicating whether the samples should be compensated prior to plotting, set to FALSE by default. If no spillover matrix is supplied to the spillover_file argument the spillover matrix will extracted from the samples. |
| spillover | name of spillover matrix csv file including .csv file
extension to apply to samples when |
| axes_trans | object of class
|
| overlay | logical indicating whether the unstained control should be
overlaid onto the plot if supplied in the flowSet, set to |
| layout | vector of grid dimensions |
| popup | logical indicating whether plots should be constructed in a pop-up window. |
| title | text to include above each plot, set to NA by default to remove titles. |
| header | vector of titles to use for the plots, set to the name of the sample by default. |
| header_text_font | font to use for header text, set to 2 by default. |
| header_text_size | text size for header, set to 1 by default. |
| header_text_col | colour for header text, set to "black" by default. |
| ... | additional arguments passed to
|
See also
Examples
library(CytoRSuiteData) # Don't run - bypass directory check for external files options("CytoRSuite_wd_check" = FALSE) # Load in compensation controls gs <- GatingSet(Compensation)#>#>#>#>#>#>#># Gate single cells using gate_draw gt <- Compensation_gatingTemplate gating(gt, gs)#>#>#>#>#>#>#># Channel match file cmfile <- system.file("extdata", "Compensation-Channels.csv", package = "CytoRSuiteData" ) # Compensation plots cyto_plot_compensation(getData(gs, "Single Cells"), channel_match = cmfile )